Downsample and crop hdf5 volumes and save as vti format¶
Download the example vistrail :
bullet-viz.vt
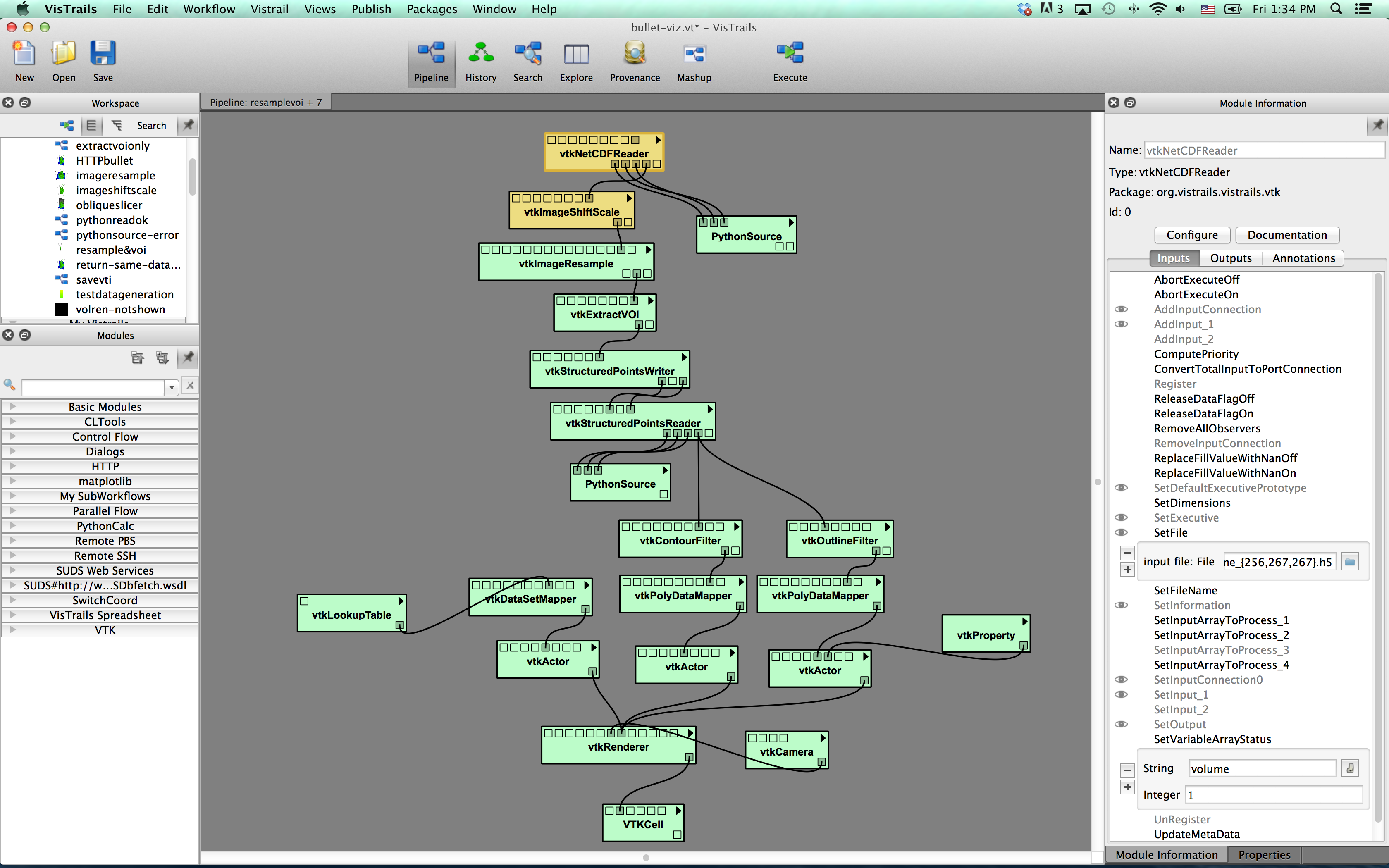
Screenshot:
Purpose of Workflow¶
- This workflow read specified dataset/variable in a hdf5 file use the “vtkNetCDFReader” module. Use a python-source module, you can write some simpe code to print out detailed information and do some test.
- The ‘vtkImageShiftScale’ module to cast the scalar type( e.g. from double to float), scale and shift the data values ( important if you cast from float to byte).
- The ‘vtkImageResample’ module allows you to interpolate the data to different axis spacing/extent with cubic, linear, or nearest neighbor algorithm.
- The ‘vtkExtractVOI’ module also can do simple subsampling, and sub-volume selection.
- The ‘vtkStructurePointsWriter’ module writes the output variable into a vtk structured point data object (usually use the file extension .vtk). The other way is to use ‘vtkXMLImageDataWriter’ module to write to vtk image data format (.vti)
- To test the new dataset, the following workflow read the new vtk( or vti, depends on which module used to write the file) file, and visualize the dataset use pseudocolor and isosurface plots.
Important modules and port setup¶
- ‘vtkNetCDFReader’ module: filename, variable array status
- ‘vtkImageShiftScale’ module: output scalar type, shift, scale
- ‘vtkImageResample’ module: Interpolation mode, AxisMagnificationFactor
- ‘vtkExtractVOI’ module: sample rate, VOI
- ‘vtkStructuredPointsWriter’ module: filename, filetype, scalar name, write
- ‘vtkXMLImageDataWriter’ module: filename, filetype, write
- ‘vtkStruecturedPointsReader’ module: filename, scalar name