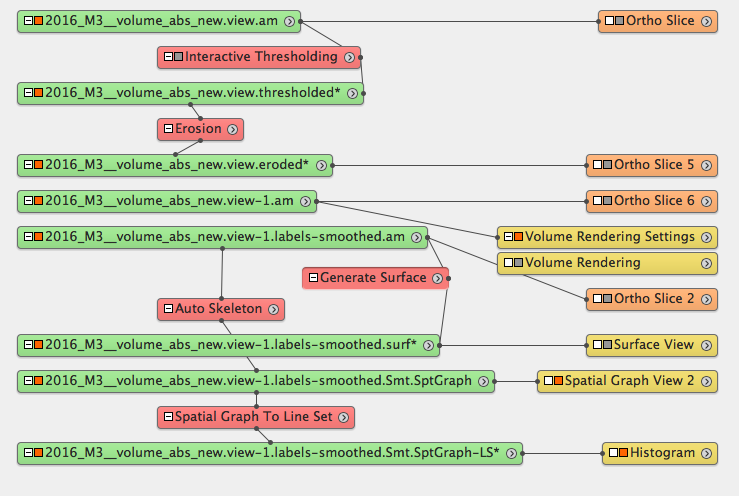
Auto skeleton¶
Dataset: 2016_M3_bin2
Crop data¶
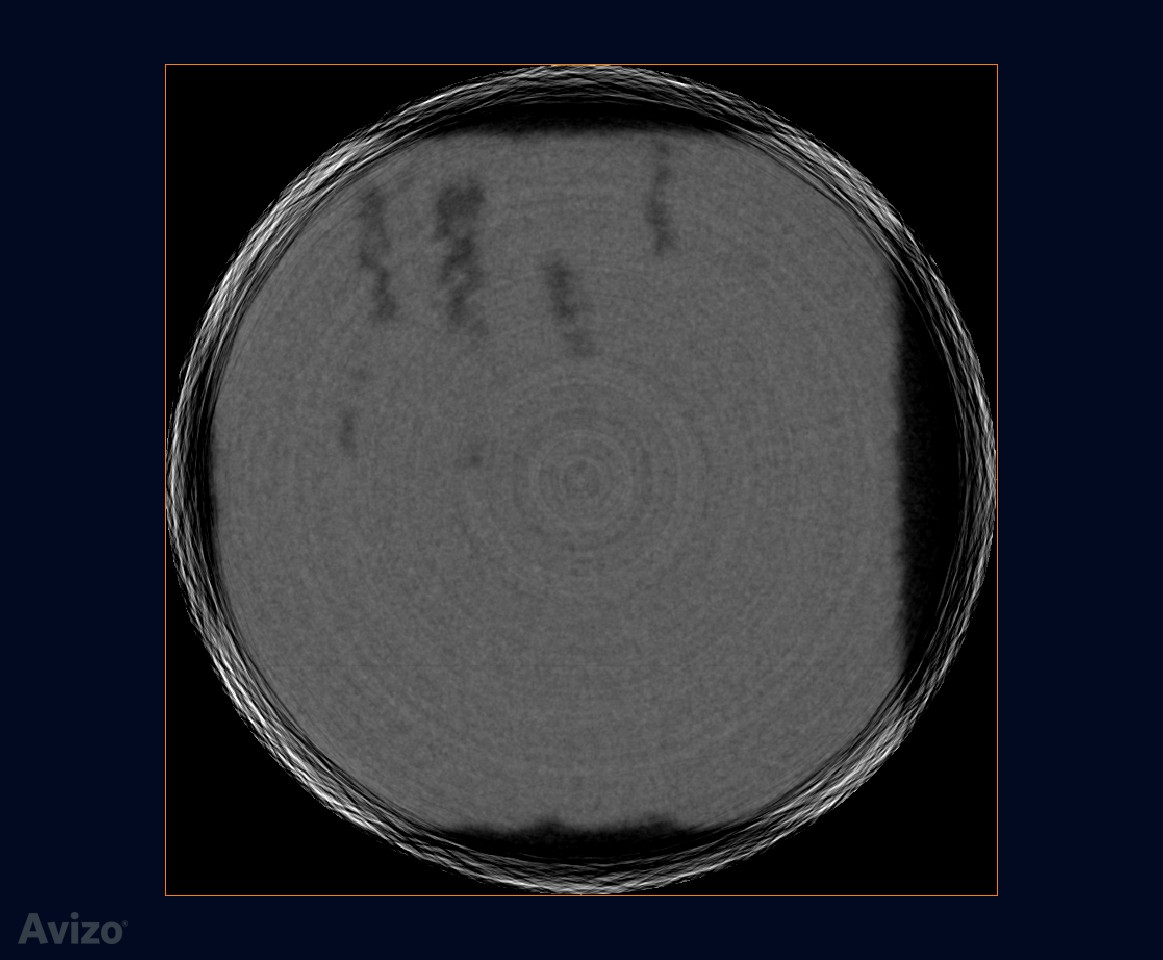
The original data is 800x800x700. At Z-direction, it’s cropped to keep index 92-654, to eliminate bad slices
Remove noise surrounding the sample¶
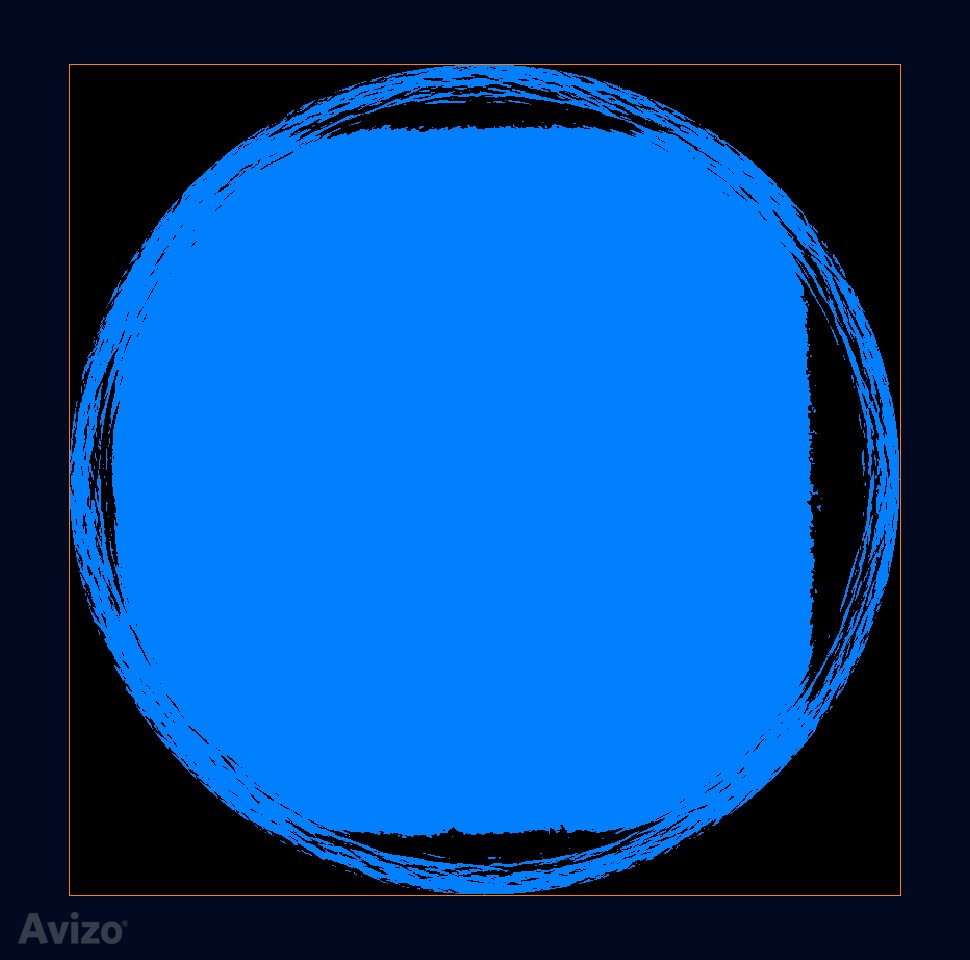
Remove noise is achieved by creating mask:
original slice
interactive thresholding
erosion: tried 3D size 5, or XY planes size 9, both works –> create a clean mask
arithmetic: data * mask –> create clean data
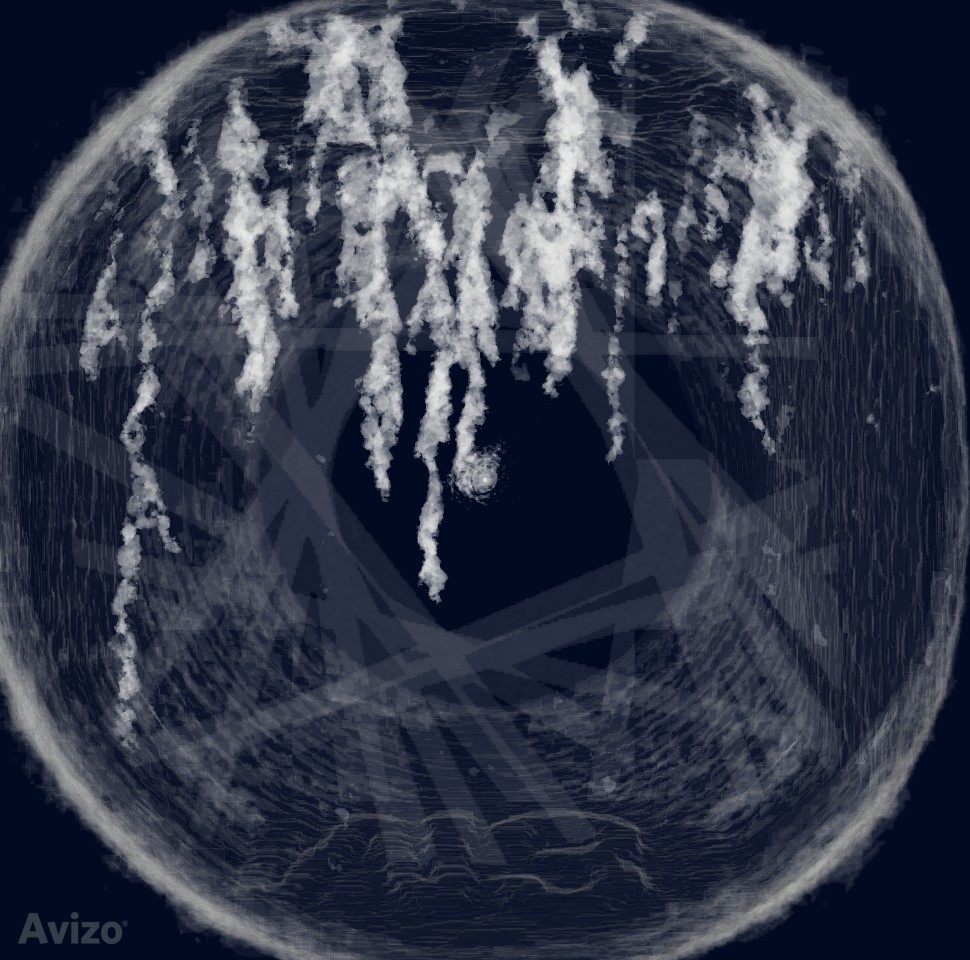
Volume rendering¶
Adjust the data window to show cracks. The important thing is to edit the colormap, so the color beyond data range is transparent.
Notice the bad slices widened after aggressive erosion
Segment the cracks¶
I used segmenation editor, magic wand + all slices. Then remove noise from menu segmentation -> smooth labels ...
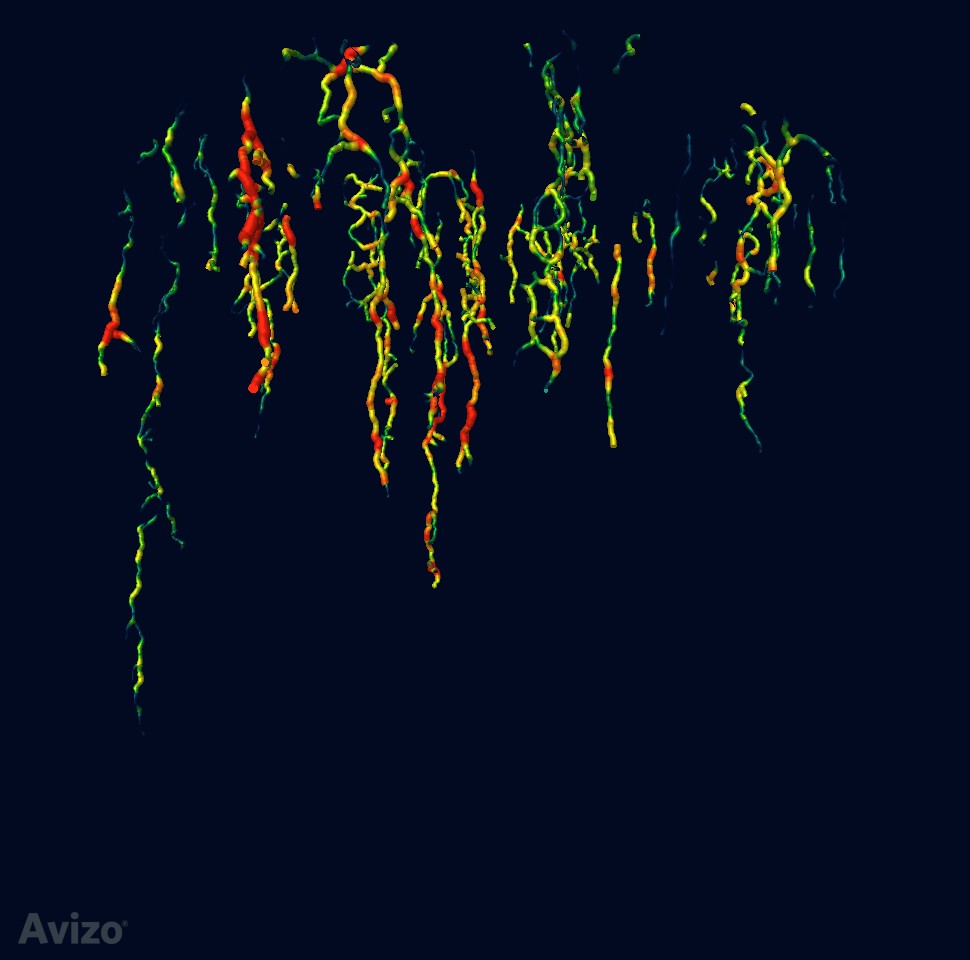
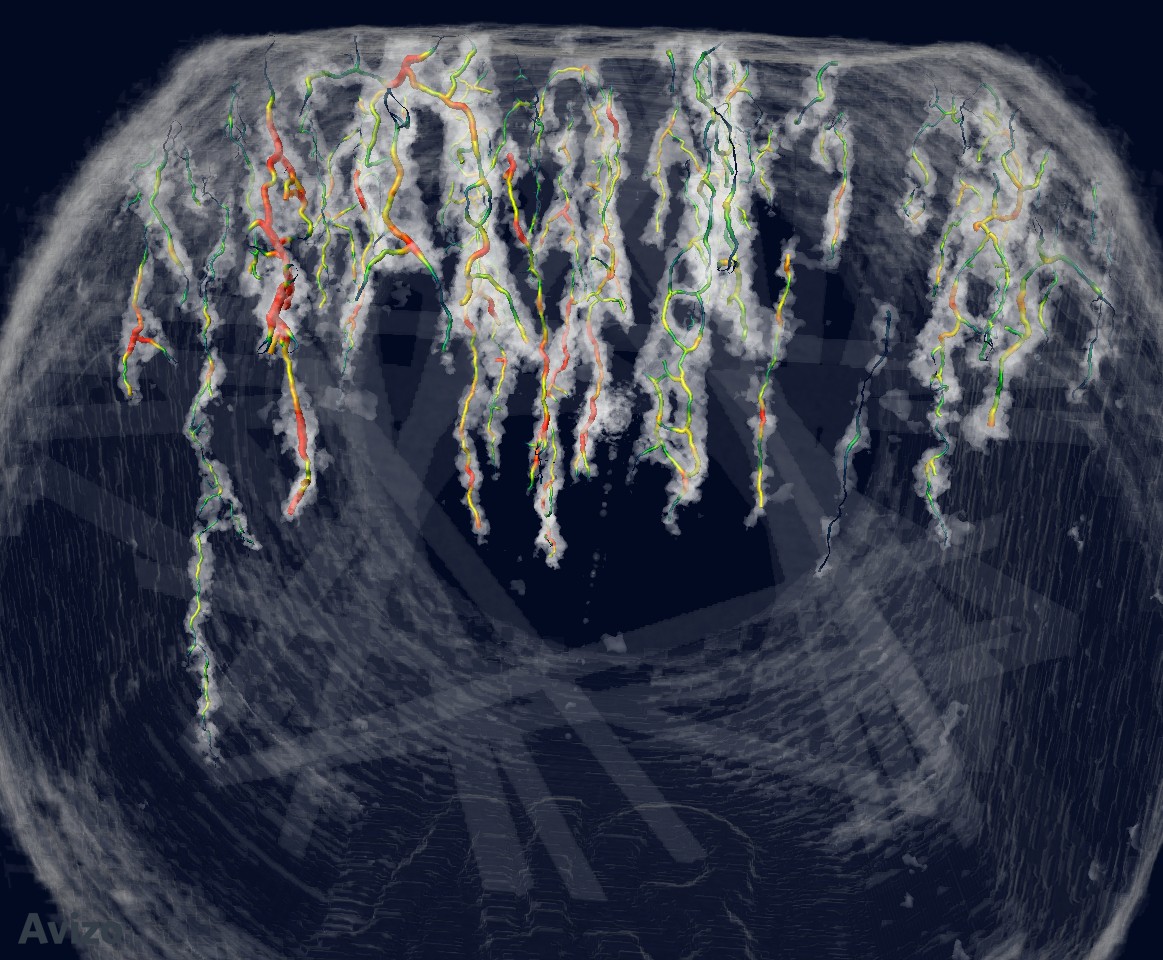
Auto Skeleton¶
Attach to crack segmentation. Use default parameters. In “Spatial graph view”, show segments, tubes, scale by thickness, color by thickness (thickness min-max: 0.5-12.1 colormap: blue to red)
Show against volume rendering:
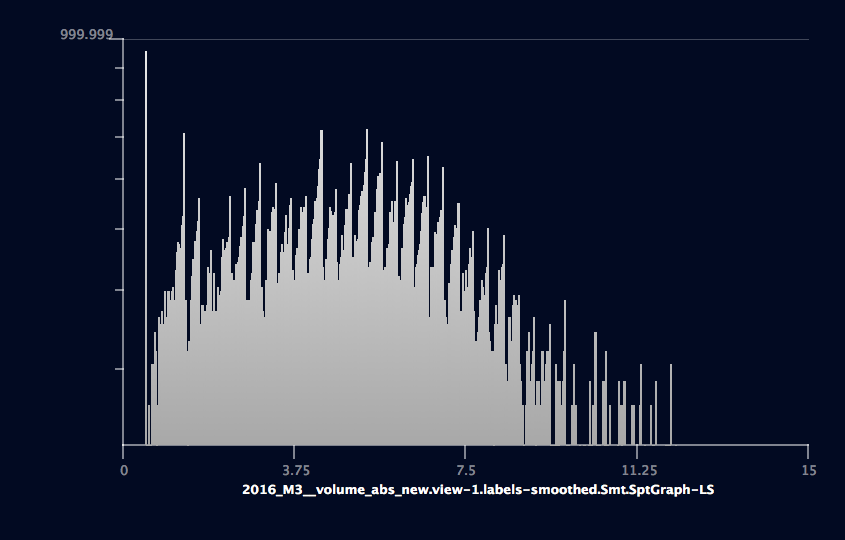
Thickness histogram¶
The SpatialGraph data can be converted to lineset. Then plot histogram on the lineset data. Choose “data 0” on “data channel” port. Click “Reset” button on “Range” port to reset data min-max. The lineset includes total 12678 points, with xyz coordinates and thickness data. The total line (segment) number is 539